We use a highly sensitive budding yeast system to collect large numbers of mutations induced by a mutagen or carcinogen of interest. Exposed single-strand DNA is roughly 100 to 1,000 times more prone to mutation than the equivalent double-strand DNA, making our system ideal for working with even weakly reactive mutagens or carcinogens.
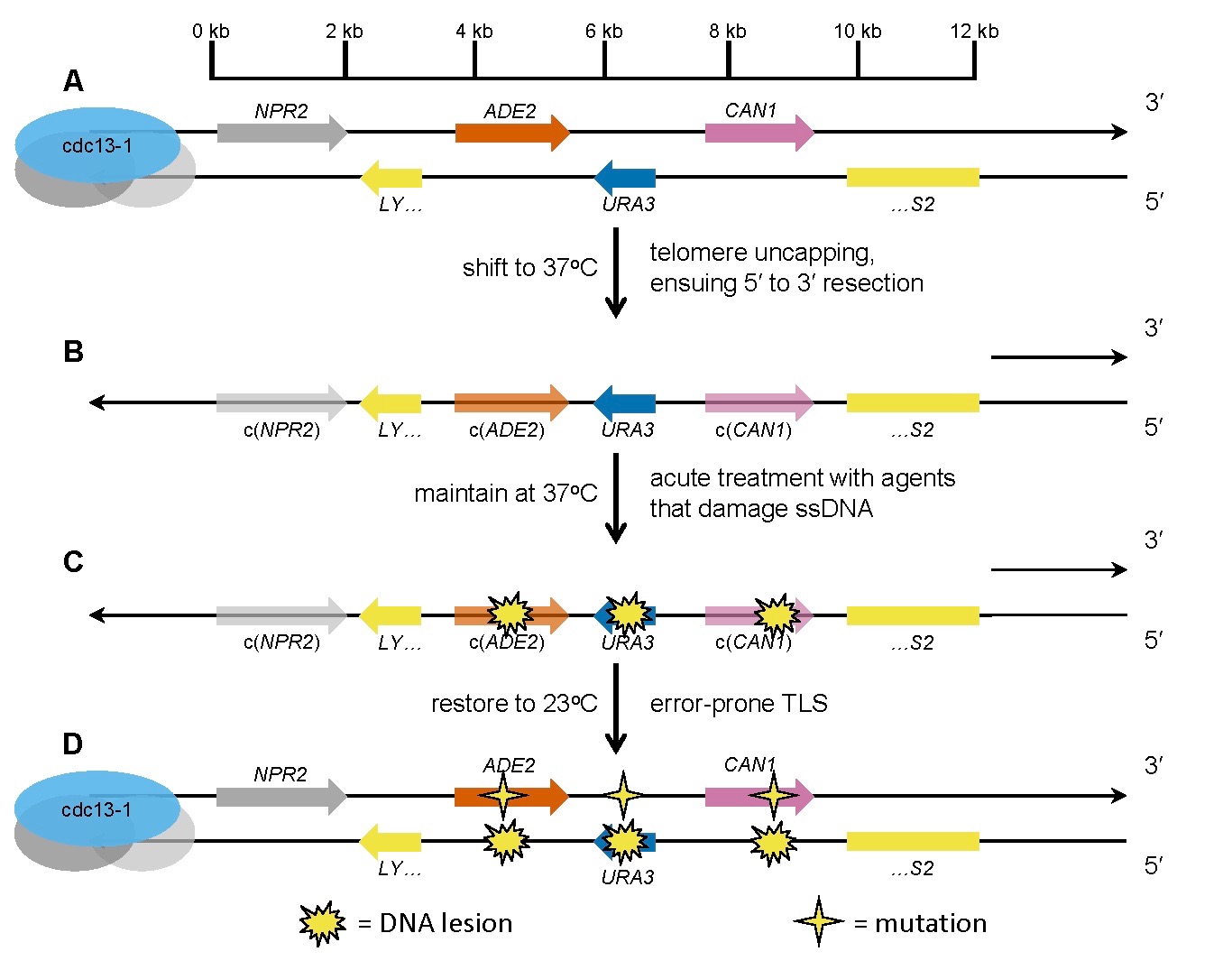
A. Three reporter genes (ADE2, URA3, and CAN1) are embedded near a telomere in a cdc13-1 haploid yeast background.
B. Shifting to 37 degrees destabilizes the Cdc13-1 mutant protein, resulting in loss of the protective proteinaceous capping structure at the telomere. The uncapped telomere is recognized as a DNA double-strand end, which is enzymatically resected to generate a long single-strand DNA (ssDNA) overhang.
C. Exposure to a test mutagen/carcinogen creates damage lesions in the exposed ssDNA.
D. Restoration to 23 degrees triggers re-synthesis of double-strand DNA. Specialized error-prone translesion synthesis (TLS) polymerases create mutations opposite the lesions. Clusters of mutations that inactivate more than one reporter gene can be selected by plating on the appropriate media.
From Chan et al., PLOS Genetics 2012.
This yeast system is sufficiently sensitive to select for mutation clusters induced by exposure to mutagens that can only damage single-strand DNA, but are not reactive enough to damage double-strand DNA.
Sulfites are used as a food preservative, including in many wines. Sulfites are not reactive enough to damage double-strand DNA appreciably, but can induce clusters of closely-spaced mutations in single-strand DNA. 30 sequenced mutation clusters induced by suflites exposure are shown here. The base positions into unique sub-telomeric DNA sequence are shown on the horizontal axis. Most mutations induced by sulfites are C (cytosine) to T (thymine) transitions. From Chan et al., PLOS Genetics 2012.
APOBEC3G is an immune system enzyme that tends to mutate the 3' C in a CC dinucleotide motif. Similar to sulfites, APOBEC3G is essentially inert toward double-strand DNA but can readily create mutation clusters in single-strand DNA. Notice however, that APOBEC3G induces a roughly equal mix of C to T transitions and C to G (guanine) transversions. This base substitution pattern recapitulates observations from cancers, where TC motif-targeting APOBEC enzymes can create many thousands of mutations per tumor. From Chan et al., PLOS Genetics 2012.
Data collected using the yeast system underpin the recent discovery than an enzyme called APOBEC3A can create tens of thousands of TC to TG and TC to TT substitutions in individual cancer samples.
a. APOBEC3A (A3A) produces a statistical enrichment for mutations at YTCA motifs (where Y = a pyrimidine base, either C or T) in the yeast system.
b. In contrast, APOBEC3B (A3B) produces a statistical enrichment for mutations at RTCA motifs (where R = a purine base, either A [adenine] or G) in the yeast system.
c. By comparing enrichment for mutations at YTCA vs. at RTCA motifs, 101 cancer genomes with an A3A-like mutation signature and 63 genomes with an A3B-like signature were identified among the 354 cancer genomes analyzed.
d. Using a more sensitive approach comparing proportions of mutations at each of the four NTCA motifs, 124 A3A-like and 75 A3B-like cancers were identified.
e. A3B-like cancers likely arose because of background mutagenic activity by A3B.
f. A3A-like cancers probably resulted from high A3A activity that overwhelms the background mutagenic signature of A3B. The molecular basis for this increased A3A activity is not fully understood.
From Chan et al., Nature Genetics 2015.
APOBEC3A-like cancers have roughly 10-fold more APOBEC signature mutations than APOBEC3B-like cancers, a finding that challenges much of the conventional wisdom in this field and could signal a major paradigm shift.
Estimated numbers of APOBEC signature TCA mutations in A3A- and A3B-like cancer samples are shown from the following types: bladder carcinoma (BLCA); breast carcinoma (BRCA); head and neck squamous cell carcinoma (HNSC); lung adenocarcinoma (LUAD); lung squamous cell carcinoma (LUSC); and all of these cohorts combined. p-values are from Kolmogorov-Smirnov tests. From Chan et al., Nature Genetics 2015.